Neil ThomasI work at the intersection of AI and biology. I am currently at EvolutionaryScale building AI tools for protein design. Previously, I was a Research Scientist at Google X. I completed my PhD in Computer Science at UC Berkeley in 2022, advised by Professor Yun S. Song. Prior to that, I was an AI Resident at Google X and a Software Engineer at 23andMe. I received my BS in Engineering Mathematics and Statistics from UC Berkeley. When I'm not being humbled by biology, I like to be humbled by a variety of hobbies. I like cooking recipes from Alison Roman, climbing rocks, skiing, playing ultimate frisbee, cycling, playing piano, watching comedy, and watering my plants. |

|
Research HighlightsMy research focuses on learning meaningful representations of proteins, with the aim of enabling applications in protein design, functional annotation, and structure prediction. Check out my thesis talk "Browsing in the Library of Babel" for an accessible introduction. |
|
Engineering highly active and diverse nuclease enzymes by combining machine learning and ultra-high-throughput screeningNeil Thomas*, David Belanger*, Chenling Xu, Hanson Lee, Kathleen Hirano, Kosuke Iwai, Vanja Polic, Kendra Nyberg, Kevin Hoff, Lucas Frenz, Charlie Emrich, Jun W Kim, Mariya Chavarha, Abi Ramanan, Jeremy Agresti, Lucy J Colwell Cell Systems, 2025 paper / code / tweetorial / talk Introduces TeleProt, a framework for guiding protein library design with machine learning that leverages evolutionary and experimental data. Validates TeleProt in an enzyme engineering campaign to optimize the endonuclease NucB. Across 4 rounds of data collection, ML-designed libraries show improved hit rate and diversity compared to directed evolution. |
|
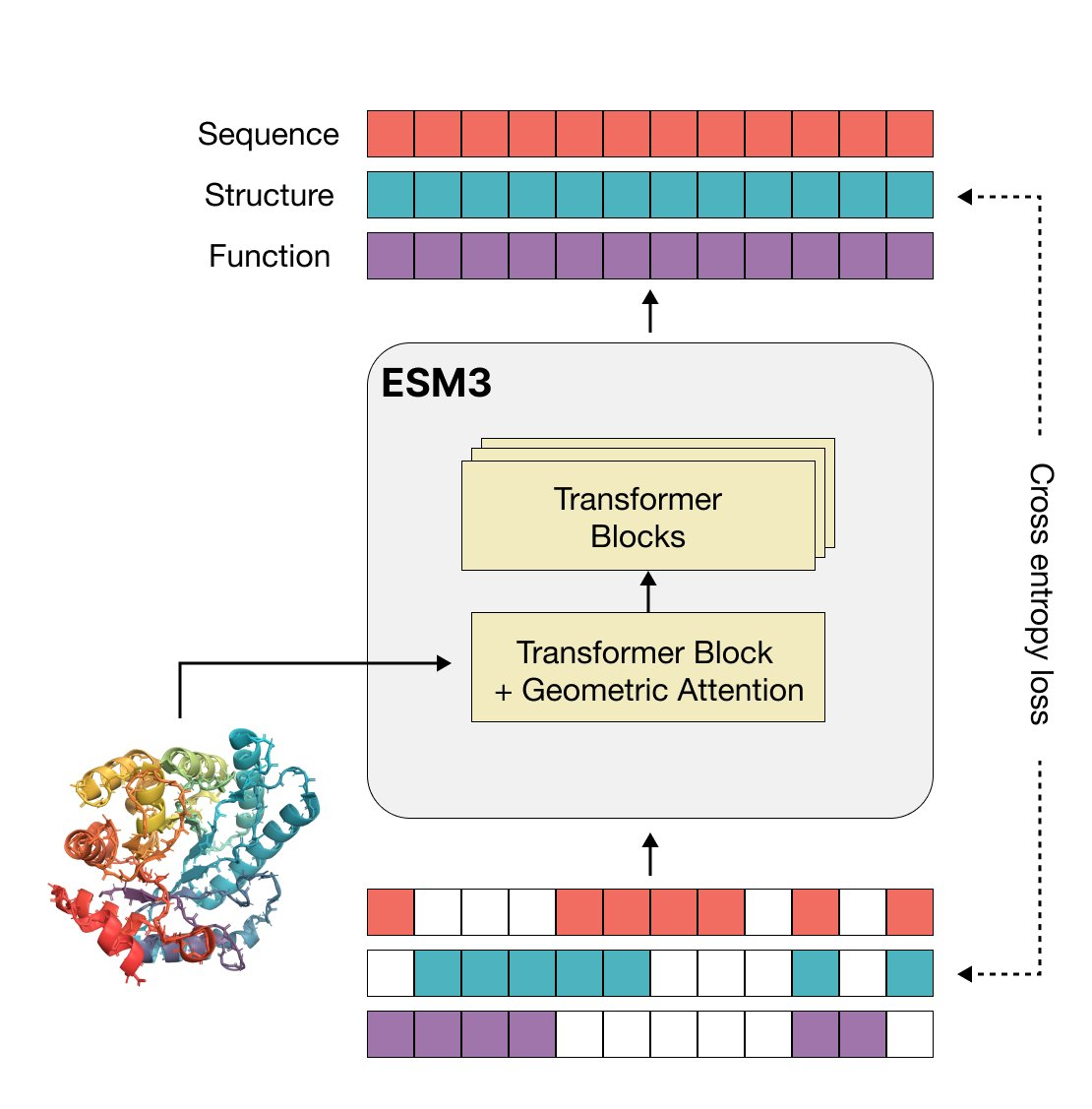
Simulating 500 million years of evolution with a language modelThomas Hayes*, Roshan Rao*, Halil Akin*, Nicholas James Sofroniew*, Deniz Oktay*, Zeming Lin*, Robert Verkuil*, Vincent Quy Tran, Jonathan Deaton, Marius Wiggert, Rohil Badkundri, Irhum Shafkat, Jun Gong, Alexander Derry, Raul Santiago Molina, Neil Thomas, Yousuf Khan, Chetan Mishra, Carolyn Kim, Liam J. Bartie, Patrick D. Hsu, Tom Sercu, Salvatore Candido, Alexander Rives Science, 2025 paper / code / tweetorial / talk / blog A multi-modal generative protein model that reasons flexibly across protein sequence, structure, and function. |
|
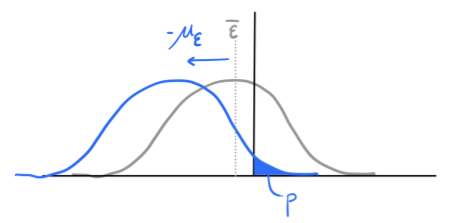
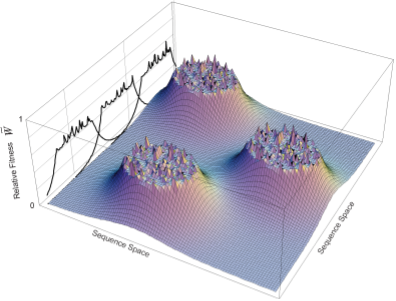
Tuned Fitness Landscapes for Benchmarking Model-Guided Protein DesignNeil Thomas*, Atish Agarwala*, David Belanger, Yun S. Song, Lucy J. Colwell bioRxiv, 2022 paper / code / tweetorial Tunable, realistic, synthetic fitness landscapes for benchmarking protein design. |
|
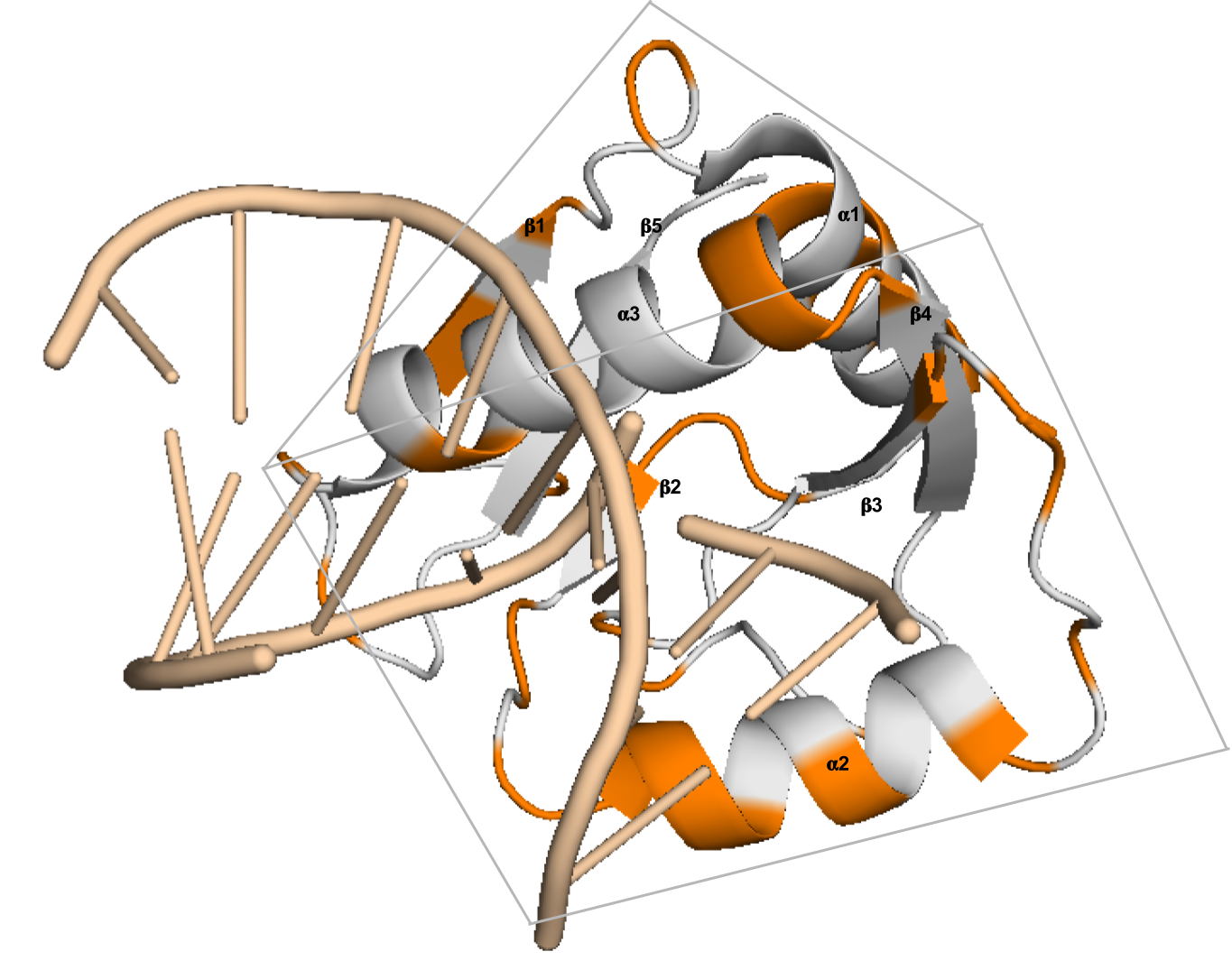
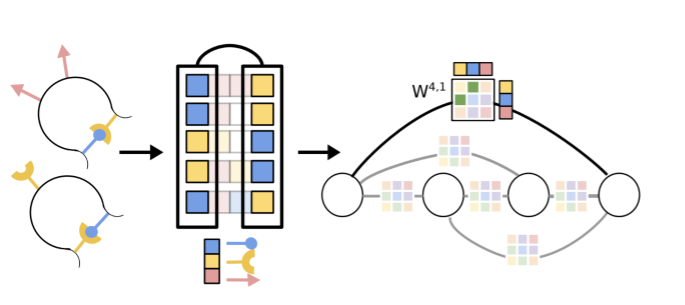
Interpreting Potts and Transformer Protein Models Through the Lens of Simplified AttentionNicholas Bhattacharya*, Neil Thomas*, Roshan Rao, Justas Dauparas, Peter K. Koo, David Baker, Yun S. Song, Sergey Ovchinnikov Pacific Symposium on Biocomputing, 2022 paper / code / tweetorial / talk Introduces “factored attention,” a simplified attention layer that we use to compare and contrast Potts models and Transformers. |
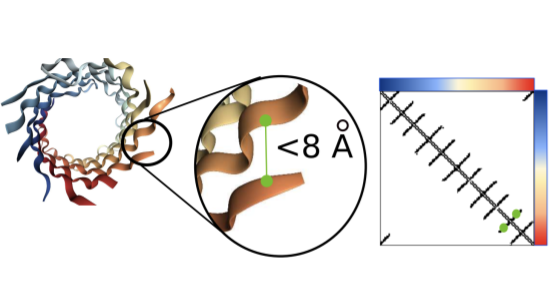
|
Evaluating Protein Transfer Learning with TAPERoshan Rao*, Nicholas Bhattacharya*, Neil Thomas*, Yan Duan, Xi Chen, John Canny, Pieter Abbeel, Yun S. Song Advances in Neural Information Processing Systems (Spotlight), 2019 paper / code / tweetorial / talk / podcast / blog A suite of benchmarking tasks for protein language models. |
ResearchFor an up-to-date list, see Google Scholar |

|
Whole-genome sequencing reveals a complex African population demographic history and signatures of local adaptationShaohua Fan, Jeffrey P. Spence, Yuanqing Feng, Matthew E.B. Hansen, Jonathan Terhorst, Marcia H. Beltrame, Alessia Ranciaro, Jibril Hirbo, William Beggs, Neil Thomas, Thomas Nyambo, Sununguko Wata Mpoloka, Gaonyadiwe George Mokone, Alfred K. Njamnshi, Charles Fokunang , Dawit Wolde Meskel, Gurja Belay, Yun S. Song, Sarah A. Tishkoff Cell, 2023 paper |
|
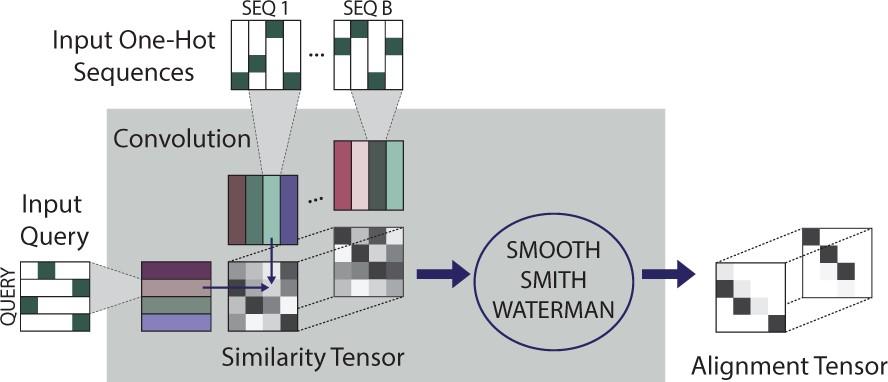
End-to-end learning of multiple sequence alignments with differentiable Smith-WatermanSamantha Petti, Nicholas Bhattacharya, Roshan Rao, Justas Dauparas, Neil Thomas, Juannan Zhou, Alexander M. Rush, Peter K. Koo, Sergey Ovchinnikov Bioinformatics, 2022 paper |
|
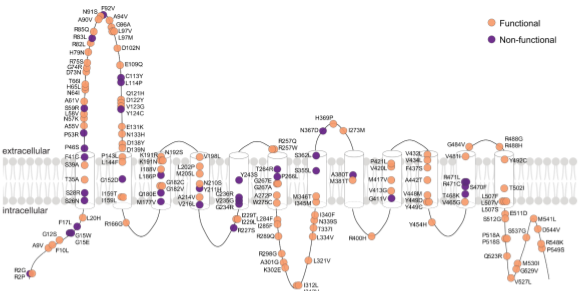
Functional genomics of OCTN2 variants informs protein-specific variant effect predictor for Carnitine Transporter DeficiencyMegan L. Koleske, Gregory McInnes, Julia E. H. Brown, Neil Thomas, Keino Hutchinson, Marcus Y. Chin, Antoine Koehl, Michelle R. Arkin, Avner Schlessinger, Renata C. Gallagher, Yun S. Song, Russ B. Altman, Kathleen M. Giacomini PNAS, 2022 paper |
|
Minding the gaps: The importance of navigating holes in protein fitness landscapesNeil Thomas, Lucy Colwell Cell Systems (Preview), 2021 paper |
TeachingDuring my graduate studies at Berkeley I had the privilege of teaching:
|
|
Built on Leonid Keselman's Jekyll fork of Jon Barron's website |